The development of Chromatin immunoprecipitation (ChIP) followed by high-throughput sequencing (ChIP-seq) tool provided a revolutionary opportunity to study different aspects of the epigenome and its interaction with the genome. ChIP-seq has been used to study different aspects of DNA packaging, histone modification, regulatory elements and the different DNA-protein interactions (outlined below). Due to the interactive nature of these aspects, ChIP-seq data can (and often is) used to query these aspects in a combinatorial fashion and, sometimes, all of them at the same time.
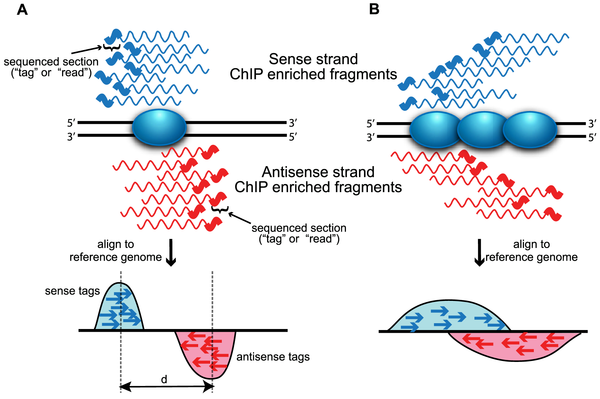
Highlights provided below showcase either the key findings or discoveries directly informing the initial aims and hypotheses. These highlights also, hopefully, emphasize the value of learning how to read ChIP-seq data.
Also Read: Is It Safe to Cook with Aluminium Cookware?
DNA Packaging, Chromatin state, and Remodeling
DNA packaging is important to ensure that DNA fits in the nucleus. DNA is packaged into nucleosomes (DNA wrapped around histones) and then into chromosomes. DNA packaging affects the accessibility of DNA, therefore affects gene expression. Genome-wide maps of chromatin states in pluripotent and differentiated murine cells gave a glimpse into the complexity of epigenetic profiles involved in genetic regulation and cell differentiation (Mikkelsen et al. 2007).
In Arabidopsis thaliana, ChIP-seq analysis of telomeres and centromeres revealed different chromatin signatures such as lower levels of heterochromatic marks (Vaquero-Sedas et al. 2012). Ting et al. (2019) found that the ubiquitin-specific protease USP11, acting as a histone deubiquitinase, is directly associated with chromatin remodeling. The role of important DNA packaging proteins, such as the linker histone (H1.0), in tumor pathogenesis and prognosis, has been studied using ChIP-seq (Torres et al. 2016). Torres et al. (2016) found that the absence of H1.0 could be linked with tumor pathogenesis and poor prognosis.
In breast cancer, the chromatin states of some of the untreated tumor cells and tumor cells unresponsive to some therapies displayed similar chromatin state signatures (Grosselin et al. 2019). These studies highlighted the importance of applications and knowing how to read ChIP seq data.
Histone Modification- Methylation and Acetylation
Histones have a wide range of states marked with different types of amino acid residue modifications, including methylation and acetylation (Wang et al. 2008, Marks et al. 2009, Peleg et al. 2010). In the human genome patterns of histone acetylation and methylation derived from CD4+ T have been studied (Wang et al. 2008). In mice, changes in acetylation of histones (e.g. histone H4 lysine 12, H4K12) have been linked with age-associated memory impairment (Peleg et al. 2010).
In murine embryonic stem cells, the Ten Eleven Translocation 1 (TET1) protein was recently identified as a partner of histone acetyltransferase Mof and scaffold protein Sin3a (Zhong et al. 2017). TET1 was also identified as a facilitator that prevents autoacetylation of the hMOF complex thus regulating expressions of downstream genes (Zhong et al. 2017).
An integrated analysis of sex determination in mice revealed epigenomic modifications such as the selective increase of H3K27me3 and a decrease in H3K4me3 in X chromosome inactivation and gene silencing across the inactive X (Marks et al. 2009). Together, these studies demonstrate how to read ChIP seq data in a way that you interpret patterns at hand to uncover key epigenomic signatures.
Identification of Regulatory Elements
Knowing how to read ChIP seq data is crucial to identifying regulatory elements and connecting their identity and function to the respective feedback networks. In embryonic stem cells, an obscure interaction of transcription factors to form large enhancer (Super-enhancers) was discovered in ChIP-seq analysis of pluripotent embryonic stem cells (Whyte et al. 2013).
Based on the activities and patterns of these Super-enhancers, the authors deduced that super-enhancers play a key role in the determination of mammalian cell fates. The epigenomic landscape of murine hearts revealed that unlike some tissue-specific enhancers, enhancers in the heart epigenome were comparably less conserved in vertebrate evolutionary history (Blow et al. 2010).
Using ChIP-seq, amongst other tools, Sigova et al. (2015) found evidence that may support that RNA plays an important role in regulating gene expression by interacting with transcription factors at gene regulatory elements.
Protein & DNA Interaction- Transcription Factor Binding
ChIP-seq analysis has shed light on DNA-protein interactions involved during development. For example, ChIP-seq analysis of mice embryonic stem cells led to the identification of precise and state-specific DNA binding motifs that recruit specific transcription factors that, in turn, determine heart development (Wamstad et al. 2012). The modulator role of the long noncoding RNA, RMST, in the interaction of SOX2 and its target genes during neurogenesis was identified through ChIP-seq analysis (Ng et al. 2013).
Modified ChIP-seq analysis of the Saccharomyces cerevisiae Rap1 transcription factor revealed binding turnover that is correlated with transcriptional output quantity (Lickwar et al. 2012). When Rap1 spent more time-bound to the genome it had higher transcriptional output whereas when it moved between binding sites and spending less time-bound per site then it had lower transcriptional outputs (Lickwar et al. 2012).
Deep learning techniques have also been explored that incorporate how to read ChIP-seq data in order to interpret and predict sequence specificities of DNA and RNA-binding proteins (Alipanahi et al. 2015).
Also Read: 3 Causes of Workplace Stress and Ways to Avoid Them
Conclusion
The major advantages of ChIP-seq also mark the governing applications of this technology, the ability to study post-translational modification and the feedback mechanisms that govern their engagement with the genome. The general theme of studies applying ChIP-seq techniques includes aims focusing on the identification of DNA packaging, histone modification, regulatory elements, and general protein-DNA interactions.
Perhaps the most valuable advantage of ChIP-seq is that it allows multifaceted interrogations of the different processes involved in gene expression and regulation that govern the very delicate and tightly controlled timing of events involved within the cells and between cells. Learning how to read ChIP-seq data opens up exciting opportunities to perform experiments designed to elucidate the world of the epigenome.